Early Career Innovators: Repurposing a Common Chemotherapy Drug for Bladder Cancer, Repurposing TIN
By Alina Shrourou, on 5 March 2021
In this Repurposing TIN interview as part of the Early Career Innovators series, acknowledging the amazing translational work being done by early career and non-tenured researchers within the UCL Therapeutic Innovation Networks (TINs), Dr Jennifer Rohn highlights her Repurposing TIN Pilot Data Fund awarded project, involving the reformulation of the common chemo drug mitomycin-C for bladder cancer.
What is the title of your project and what does it involve?
My project is entitled “Repurposing mitomycin-C to expand opportunities for bladder cancer sufferers”. We decided to take advantage of a previous therapy platform we developed for urinary tract infection (UTI) to see if we could retrofit it for bladder cancer therapy with a few tweaks.
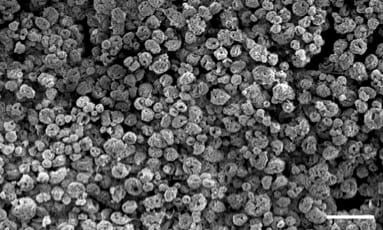
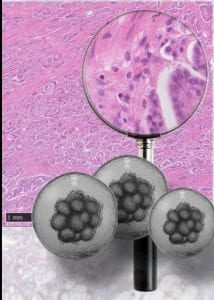
Our UTI solution, which is currently being commercialised by the UCL spinoff AtoCap in collaboration with engineering colleagues at UCL and Oxford, is a reformulation of the common antibiotic nitrofurantoin. The antibiotic is mixed with the FDA-approved polymer PLGA using patented technology, resulting in novel microcapsules that are designed to be delivered directly into the bladder via catheter, and once there, to penetrate robustly, delivering drug deep into the bladder wall where it’s needed. In this TIN award project, we are going to reformulate the common chemo drug mitomycin-C using the same platform, in the hopes that the resulting microcapsules will allow deep bladder delivery of this important drug which is currently of limited use because it’s difficult to get it into the tissues.
In parallel, in order to test and troubleshoot our prototype chemo-capsules, we are adjusting a human bladder “organoid” model developed in my lab so that it is equipped with fluorescent cancer cells. Using this modified platform as a test-bed, we should be able to do some great experiments with the prototype capsules to see if they can penetrate and kill cancer cells better than free chemo drug. This data package will be used to attract follow-on funding to help speed our solution down the translational and commercial pathway.
What is the motivation behind your project/therapeutic?
Bladder cancer is a bit of a neglected disease; it’s the most expensive cancer of all to treat, but unlike other cancers, outcomes have not improved for decades because there hasn’t been much research into mechanisms and new drug discovery. We are focusing here on non-muscle-invasive bladder cancer (NMIBC). Of 300,000 new NMIBC cases per year worldwide, low/intermediate-risk patients (~80%) would benefit from more penetrative chemotherapy, and it might newly enable treatment of high-risk forms and relapsed patients. The current standard of care is surgery, followed by traditional chemotherapy and Bacillus Calmette–Guérin (BCG). Recent advances in immunotherapy, while promising, are suitable for only a limited number of advanced cases, and are associated with severe side effects. Patient support groups repeatedly identify improved NMIBC treatments as the top priority.
Can you highlight any challenges have you experienced as an early career researcher in the repurposing/translational research space?
Although I’ve been a scientist for a long time, I only recently started up a lab of my own due to my convoluted career pathway in and out of academia. Just staying afloat in academia as “a mature ECR” is a challenge in itself, and though I still don’t have a permanent position, I am clinging on in there. I think reformulation is particularly tricky. Although the route-change offers a streamlined regulatory pathway, which lowers costs, it is sometimes difficult to persuade investors that a generic drug can be reinvented into something new and profitable, and your patent position has to be really solid. It’s been interesting to be involved in a university spin-off – I was recently made Chief Scientific Officer, so I get exposed to a lot of the business side as well as the science. It’s a truly fascinating world and I am still learning all the lingo!
Why did you want to apply to the Repurposing TIN Pilot Data Fund?
I’ve been involved in the TIN for a while and it seemed a natural fit for my project. We need pilot data to get further funding, but you can’t do research without money. The TINs Pilot Data Fund therefore was a really attractive way to get around that “chicken and egg” problem.
How did you find the process for the TIN Pilot Data Fund? What did you learn?
I think the whole thing was beautifully organised and I particularly valued the professional pitch training we received. It not only helped me win the award, but I can use these skills to help improve my pitching to various investors. The organisers were also so helpful throughout the process.
Translational training from UCL ACCELERATE
What do you hope to achieve in the duration of your project?
Things have been very challenging because of COVID and even though the labs are now open, everything is going a lot more slowly due to ongoing lab occupancy restrictions. Thankfully the scheme has been extended for those experiencing issues due to restrictions of the pandemic, to give us all a little more breathing space. Things are going very well so far – we are nearly ready to merge our fluorescent cancer cells with our healthy bladder “organoid” and we expect to have our data package completed right on schedule.
About Dr Jennifer Rohn
Dr Jennifer Rohn, a cell biologist, is Principal Research Associate and Head of the Centre for Urological Biology in the Department of Renal Medicine in the Division of Medicine, based at the Royal Free Hospital campus.
After receiving her PhD from the University of Washington in Seattle studying virus evolution, she held several post-doctoral positions in academia and industry in the Netherlands and in the UK (along with a research career break in science publishing) before settling into her current role. She and her research team are interested in understanding urinary tract infection and as part of this, they seek to find novel therapeutic delivery mechanisms that facilitate penetration of the bladder wall to kill bacteria sheltering within. More recently the lab has branched out into bladder cancer, another disease in need of better penetrative solutions for chemotherapy. Jennifer is also a prolific writer and science communicator in her spare time, and she has published three novels about scientists (a genre known as “lab lit”).
 Close
Close