In this Devices & Diagnostics TIN interview as part of the Early Career Innovators series, acknowledging the amazing translational work being done by early career researchers within the UCL Therapeutic Innovation Networks (TINs), Dr Nikolas Pontikos highlights his Devices & Diagnostics TIN (co-lead by the UCL Institute of Healthcare Engineering’s Translational & Industry Delivery Group) Pilot Data Fund awarded project, involving the use of artificial intelligence to accelerate genetic diagnosis of inherited retinal disease.
What is the title of your project and what does it involve?
The title of my project is: “Eye2Gene: Accelerating Genetic Diagnosis of Inherited Retinal Disease with AI”
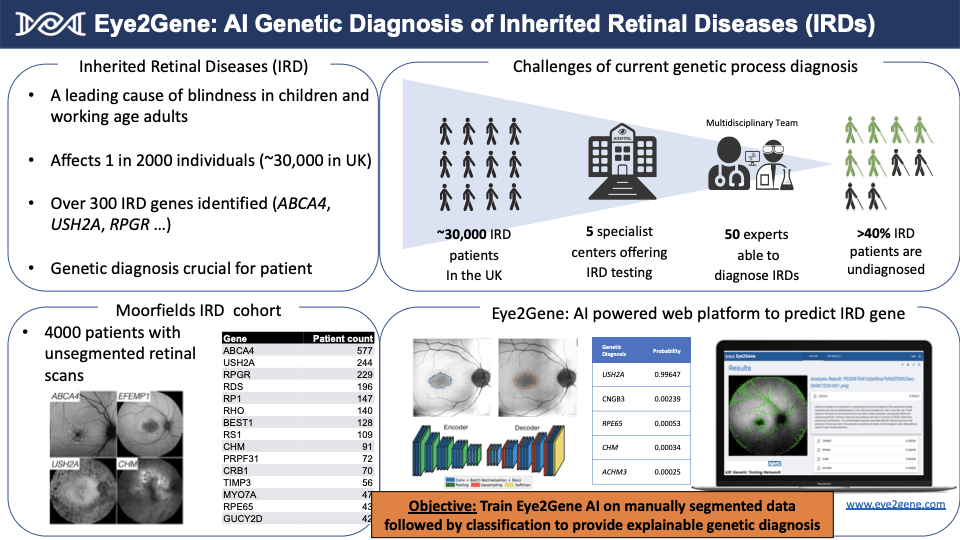
Inherited retinal diseases are a leading cause of visual impairment in children and the working age population. Mutations in over 300 genes are associated with IRDs and identifying the affected gene in a patient is the first step towards diagnosis, prognosis and treatment. Currently, inherited retinal diseases are detected first by retinal imaging analysis and later confirmed by genetic analysis. Teams combining these analytical skills are scarce hence my idea is to train an AI (artificial intelligence), Eye2Gene, to achieve this in one algorithm. The training data will consist of retinal images from 4000 inherited retinal disease cases at Moorfields Hospital segmented over a 2 month period.
What is the motivation behind your project/therapeutic?
Around 30,000 individuals in the UK have an inherited retinal disease (3.5M globally). Less than 40% of patients have been diagnosed because of poor screening. A late diagnosis means less chances for treatment. Genetic diagnosis is crucial for management and treatment of patients by upcoming gene-targeted treatments. Rare disease drug development is one of the fastest growing pharma markets ($262bn by 2024). Eye2Gene will increase the rate of genetic diagnosis, allowing more to be treated sooner. There are currently no competing products for inherited retinal disease genetic diagnosis.
Why did you want to apply to the Devices & Diagnostics TIN Pilot Data Fund?
I first heard of the Translational Innovation Network Pilot Data Fund through the UCL newsletters of Personalised Medicine. I attended a few events organised by the UCL Translation Research Office and was really impressed by the guidance and support that was provided to Early Career Researchers such as translational pathways, presentation skills and grant writing workshops often led by experienced professional external consultants.
Having recently published the dataset for inherited retinal diseases from Moorfields Eye Hospital (Pontikos et al., 2020) and developed the deep-learning algorithm Eye2Gene prototype, the Pilot Data Fund seemed like the ideal kickstarter grant to launch my research project in order to build a pilot imaging dataset of segmented inherited retinal disease scans to allow for explainable AI and enhance algorithm performance.
How did you find the process for the TIN Pilot Data Fund? What did you learn?
The process was very educational. The workshops organised were of a very high standard and for the first time in my career, I received professional training in writing grants and pitching ideas. I also learnt about the translational pathway and the different stages of technology readiness. I think perhaps two workshops that stood out for me were the ones presented by Granted Ltd and by Simon Cain. As an exuberant scientist, project management (Gantt charts, KPIs, risk management etc) is always something that came as an afterthought for me, so it was quite revealing for me to see just how central it is to the grant writing process and funding success. In the future, through additional TIN opportunities, I am also very much looking forward to learning more about regulatory aspects of medical device development.
What do you hope to achieve in the 6 months duration of your project?
After 6 months I hope to have a dedicated retinal imaging annotation platform and a large manually segmented dataset of inherited retinal disease scans (>2000). This will allow Eye2Gene to offer interpretable output, highlighting to healthcare professionals exactly which parts of an image were used to derive a diagnosis. These outputs will be useful for the training of multiple AI algorithms including Eye2Gene.
Furthermore, the retinal image annotation platform that will be developed will support future image segmentation projects by facilitating collaborative editing and training of medical graders, as well as supporting medical image annotation for clinical trials. In the future, I hope to share these annotated rare disease datasets with the community and promote natural history studies and drug development. On the back of this funding from TIN, I have already submitted a project grant application to NHSX and the Wellcome Trust to support further development of the Eye2Gene software as a medical device.
What are your next steps from now?
For the Eye2Gene project I have already exported the inherited retinal disease imaging datasets and the software development of the image annotation platform has started which should be finished by end of May. After which, the annotation of images will start and I anticipate will finish in July. Further to this, the UCL Translational Office has been incredibly supportive in helping me apply for large project grants such as the Wellcome Trust Innovator Award and the NHSX AI Award. They have helped me connect with relevant individuals outside of my area of expertise, such as experienced project managers, regulatory consultants and health economists at UCL. These individuals have taken an active part in helping me write my grant applications which has been really fantastic! I will hear back from these grants in February and hope to be successful (fingers-crossed).
Whether or not I am successful with these grant applications in the short-term, I believe the whole process has greatly strengthened my grant and fellowship writing skills, especially by teaching me good project management and pitching skills.
Career wise I have also recently submitted two fellowship applications one to NIHR and one to MRC which if I am successful, will start in October 2021, giving me five years of funding. I also plan on submitting a studentship to hire a PhD student (as subsidiary supervisor).
About Dr Nikolas Pontikos
Dr Pontikos is an early career researcher funded by a short-term Moorfields Eye Charity Career Development Award. He is based at the UCL Institute of Ophthalmology and Moorfields Eye Hospital, and collaborates with the Institute of Health Informatics and the Genetics Institute. He has an MEng in computer science from UCL, a postgraduate MSci in bioinformatics from Imperial College and a PhD in genetics and machine learning from Cambridge University. He is very interested in the analysis of healthcare data to provide personalised care.
He jointly analyses genetics, medical imaging and text data to develop decision support systems for diagnosis, prognosis and treatment. His focus has mostly been on rare eye diseases but his methodology is widely applicable to rare genetic diseases. He is very interested in learning more about the regulatory aspects of developing software as a medical device.
References
Pontikos, N., et al. (2020). Genetic basis of inherited retinal disease in a molecularly characterised cohort of over 3000 families from the United Kingdom. Ophthalmology. https://doi.org/10.1016/j.ophtha.2020.04.008
 Close
Close




