Male Promiscuity Boosts Role of Chance in Sex Chromosome Evolution
By Claire Asher, on 19 March 2015
Humans, like all mammals and birds, determine sex with chromosomes. Whether a fertilised egg develops into a male or female depends on what chromosomes it carries Scientists have long recognised that genes evolve a little differently on the sex chromosomes, and recent research in GEE suggests this may be due to differing patterns of inheritance that favour the influence of chance on gene sequence change. Furthermore, promiscuity in males has a large influence on the magnitude of this effect, with chance playing an even greater role in sex-chromosome evolution in highly promiscuous species. Using genetic sequence data in combination with physical and behavioural measurements of promiscuity in birds, Dr Alison Wright and Professor Judith Mank report strong evidence for the role of neutral forces in sex chromosome evolution.
In birds and mammals, along with some invertebrates and reptiles, sex is determined by the chromosomes you carry – the sex chromosomes, as they are aptly named. If you are a male mammal, you carry one X and one Y chromosome; a female mammal carries two X chromosomes. Similarly, if you are a male bird, you carry two Z chromosomes; a female carries one Z and one W. Whether it’s the XY system, the ZW system or even the UV system used by some species of algae, the result is more or less the same. Sex is determined by the presence or absence of particular chromosomes. This isn’t always the case – some species determine sex using temperature during development, other species determine sex based on social conditions, while others do away with fixed sexes altogether and are either hermaphrodite or possess the ability to switch sex. However, one of the most common, and certainly the best studied, systems among living organisms is to determine sex with chromosomes.
Unlike autosomal chromosomes (all our chromosomes that are not sex chromosomes), sex chromosomes are not inherited and expressed equally across the sexes. The Y and W chromosomes only ever appear in one sex, for example. This has some interesting consequences for evolution. For example, scientists have found that the ‘major sex chromosomes’ (X and Z chromosomes) tend to evolve faster than the autosomes. Known as the Faster-X (or Faster-Z) effect, this phenomenon is now well documented in a range of different species, and scientists have suggested a number of possible explanations for why this might be the case. Faster evolution on the major sex chromosomes might be caused by more effective natural selection favouring beneficial mutations (adaptive hypothesis) or due to less effective natural selection failing to remove harmful mutations (neutral hypothesis).
Why would natural selection act differently on sex chromosomes than autosomal ones? In a paper published in Molecular Ecology this month, Dr Alison Wright explains that the differences between chromosomes arise because of differences in the pattern of inheritance, which ultimately influences the number of chromosomes that are passed on to the next generation, called the effective population size. An individual who never reproduces is an evolutionary dead end, and as their genes are not passed on, and they are not counted in the effective population size. Individuals that do mate contribute sex chromosomes unevenly, and this can have a significant impact on the course of sex-chromosome evolution.
When two individuals mate, they each pass one of each pair of chromosomes to the offspring. Each chromosome has an equal likelihood of being carried by the offspring, and the effective population size (ie chance of being passed on) of all autosomal chromosomes is the same. But for the sex chromosomes, things are a bit more complicated. Each time a pair of individuals mate, between them they bring three major sex chromosomes and one minor chromosome to the table. This translates to major sex chromosomes having an effective population size three times larger than the minor sex chromosomes. And both sex chromosomes have a smaller effective population size than the autosomes.
But that’s only if everybody is monogamous. As soon as promiscuity is involved, things get even more complicated. If males are promiscuous (and they often are, in the animal kingdom), then this means some males in the population are likely to be very successful, while others fail to reproduce at all. In other words, the variance in male mating success is much higher. Promiscuity reduces the effective population size of the minor chromosomes even further.

Why does effective population size matter? Well, the effective population size determines the relative influence of chance on gene sequence evolution. Although we generally think of evolution progressing as natural selection favours beneficial mutations and purges deleterious ones, chance also has a big role to play. Chance, known in this context as genetic drift, has a bigger impact on small populations, and rare mutations. This is because when a particular mutation is rare, it only takes a little bit of bad luck for it to be lost forever. Just think of the times you’ve walked home in the rain only to hear the characteristic crunch of the end of a snail’s life – here your foot is the agent of genetic drift. The death of that snail had little or nothing to do with the genes it carried, but your foot has altered the course of evolution, slightly. The effective population size of autosomal genes reflects the population size of the organisms they are found in, but for the sex chromosomes, their effective population size is even smaller, making them more prone to genetic drift.
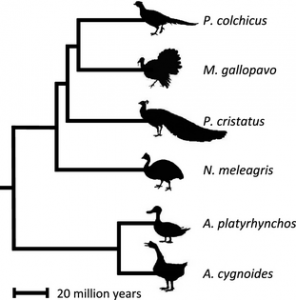
Dr Alison Wright, Professor Judith Mank and colleagues from GEE sequenced expressed genes in six species of birds, spanning 90 million years of evolution, to investigate the rate of evolutionary change in genes on different chromosomes. They compared sequence data from monogamous species like the Swan Goose (Anser cygnoides) and the Guinea Fowl (Numida meleagris) with promiscuous species like the Mallard duck (Anas platyrhynchos), wild Turkey (Meleagris gallopavo), and Peafowl (Pavo cristatus) to investigate how gene sequences and gene expression patterns vary both within and between species. They then matched data on the rate of evolution with characteristics of species that are associated with promiscuity, such as testes weight and sperm number. Their results indicate that natural selection is less effective on the Z chromosome in general, and this becomes even more pronounced in promiscuous species. The authors therefore conclude that Faster-Z evolution in birds is not adaptive, but is driven by neutral processes.
Differences in gene sequences within and between species can tell us a lot about the rate of evolution for different lineages. This is because the genetic code has some redundancy in it – DNA is split up into three-letter words or codons, and there are many cases where different codons translate into the same amino acid. So, it is possible to have genetic sequence change that is essentially invisible to natural selection – it doesn’t alter the resulting protein sequence and so has no influence on the organism. Changes in gene sequence that swap between these ‘synonymous’ codons can therefore give us a rough baseline of neutral change. Non-synonymous differences (the ones that do have an effect on the organism), between individuals or between species, represent the rate of evolution. More non-synonymous changes suggests either positive selection, where evolution favours those changes because they are beneficial, or genetic drift, where selection is weaker and cannot remove slightly harmful mutations from the population. The authors found that genes on the Z chromosome show a faster rate of non-synonymous change than autosomal genes. Further, the ratio was significantly correlated with measures of promiscuity, with more promiscuous species having more non-synonymous changes.
Although this could be a mark of positive natural selection, the authors found no difference in the number of genes undergoing positive selection between sex- and autosomal-chromosomes, suggesting the Faster-Z effect is driven by genetic drift rather than positive selection. In fact, differences within species indicate that natural selection is less effective at removing mildly deleterious mutations from the Z chromosome than the autosomes. Combined with other analyses on gene expression, these results show strong support for the neutral explanation for Faster-Z evolution in birds.
Interesting, promiscuity increases the effective population size of X chromosomes, and that may explain why previous studies have found evidence that Faster-X chromosome may well be due to positive selection. These differences suggest that Z chromosomes may be less important in adaptation than X chromosomes.
Original Article:
This research was made possible by funding from the Natural Environment Research Council (NERC) and the European Research Council (ERC).
 Close
Close