In the first Small Molecules TIN interview as part of the Early Career Innovators series, acknowledging the amazing translational work being done by early career researchers within the UCL Therapeutic Innovation Networks (TINs), Clara Gathmann highlights her Small Molecules TIN Pilot Data Fund awarded project, “Screening a DNA encoded library on GEF-H1 for drugs targeting ocular diseases”.
What is the title of your project and what does it involve?
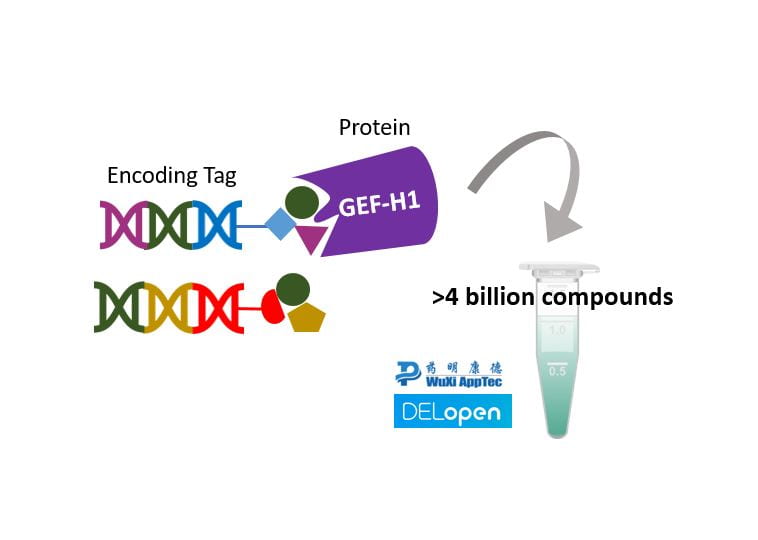
The title of my project is ‘’Screening a DNA encoded library on GEF-H1 for drugs targeting ocular diseases’’. GEF-H1 is a protein which our group has identified as a potential target for fibrotic and inflammatory ocular disorders. We have started to develop small molecules against GEF-H1, using computational and medicinal chemistry drug discovery techniques. However, our molecules are not very potent yet and we haven’t tried using high throughput screening.
DNA encoded libraries (DELs) contain billions of molecules in a single tube and can be screened on a protein within days. This technology is made possible by DNA tags attached to each molecule that encode their structures. As you may know, DNA can be amplified with PCR and sequenced. This enables a read-out of the structures of the most active molecules from picogram quantities in the tube. Hence, only one small tube is needed instead of thousands of 96-well plates. We decided to use the open source DEL library from WuXi and subject it to GEF-H1. With this, we hope to discover new molecules that bind GEF-H1 within a few weeks, giving a real kick to our drug discovery plans!
What is the motivation behind your project/therapeutic?
Preventable ocular disorders are still a major cause for vision loss. In addition to the high impact on human lives, sight loss is also a real economic issue for our societies. Even quite common ocular disorders like uveitis and retinopathies can cause vision loss if left untreated. Unfortunately, current treatments for inflammatory and fibrotic eye disorders either involve invasive surgeries or the heavy use of drugs like corticosteroids and anti-proliferative agents. These interventions are first of all not necessarily successful and have several side effects, including even vision loss!
As for many diseases, it all starts with a good drug target (in other words, a protein to inhibit). By finding potent GEF-H1 inhibitors, we hope first of all to produce useful clinical candidates that can prevent inflammatory and fibrotic damages done to the eye. But also importantly, we would show for the first time that GEF-H1 is a suitable protein to inhibit, paving the path for a new class of biological targets and providing a proof of concept for future drug discovery projects. I think this is truly motivating, to not only contribute to a cause such as a particular disorder, but also to a whole scientific field which might help completely unrelated disease classes.

Why did you want to apply to the Small Molecules TIN Pilot Data Fund?
DNA encoded libraries are an emerging technology that have gained a lot of attention lately. More and more data is published on the construction of such libraries, with some successful examples taken to clinical trials. However, the biggest libraries are often in-house libraries of pharmaceutical companies or offered by specialised companies to pharma giants. This means that drug discovery groups in academia don’t have easy access to these libraries for cost reasons, unless a collaboration is put in place. When we heard that WuXi launched this open source DNA encoded library for academia, it sounded like a huge opportunity to bring that technology into UCL and our department.
This technology can lead very quickly to positive results but would simply not have been possible to follow up on hits without the funds from the Small molecules TIN. The TIN pilot data fund seemed to be adapted due to the short-term character of the project and we hoped that such an unusual idea could awaken interest. In addition to this, applying for this kind of fund seemed like the perfect opportunity for me to learn about grant writing and how to fund research in general.
Learn more about the Therapeutic Innovation Networks and join a TIN
How did you find the process for the TIN Pilot Data Fund? What did you learn?
Overall, super exciting. While making the written application, I realised how important it is to connect our (sometimes crazy) scientific ideas to real life goals. For the first time in my life, I had to define the purpose for what I am doing in such details. In some disciplines like the ones involving clinical research, it might be easier to relate to the patients, but when you are in a lab synthesising molecules, sometimes you just loose that connection. Applying for the fund made me realise that the translational aspect of research even at its early point is really important.
Then, there was the pitching, which felt a bit like preparing for a TV show! We candidates had the chance to participate to an ACCELERATE workshop on pitching, probably the most important part of the application process. My pitch before and after that session was transformed thanks to the honest comments I received. I learnt to shift completely the initial ‘science nerdy’ focus of the talk to ‘why you should fund my project’. In summary, I learnt to tell why my project is going to make a change, and how I am going to achieve my goals in time.
What do you hope to achieve in the 6 months duration of your project?
The goal for this project is mainly to generate molecules that can be useful biological tools or clinical candidates which target GEF-H1 in ocular diseases. We want to generate accurate binding data on the hit molecules to prepare them for in vivo testing. If the screen generates potent molecules that bind GEF-H1 tightly, this will enable us to irrevocably confirm that GEF-H1 inhibition is beneficial for those diseases, fast-tracking us to clinical testing.
What are your next steps from now?
In those six months, I will first prepare materials for the screen (immobilise the proteins on beads for example), to then subject the protein to the DEL screen. After the screen is done, WuXi will process our samples, amplify the DNA tags and perform a statistical analysis on these to reveal the structures of the binders. We will then order the most potent binders for resynthesis on a milligram scale and validate them using biological assays. We typically perform biophysical assays like surface plasmon resonance (SPR) and cellular assays that model for example inflammation.
About Clara Gathmann

Clara Gathmann works between the UCL Institute of Ophthalmology and the Wolfson Institute for Biomedical Research. She is working in the groups of Prof. Balda/Matter and Dr.Chan/Prof. Selwood, focussing on the discovery of small molecules as drug candidates for common ocular diseases. She started on a Moorfields Eye Charity funded project in October 2019 involving the design, synthesis and biological assessment of molecules inhibit the GEF-H1/RhoA interaction.
 Close
Close










