Lunch Hour Lecture: Bones, mummies, tuberculosis and ancient DNA
By Ella Richards, on 18 March 2016
As World Tuberculosis Day approaches on 24 March, Dr Helen Donoghue (UCL Clinical Microbiology) ends this term’s series of Lunch Hour Lectures by looking back at 9,000 year old tuberculosis DNA.
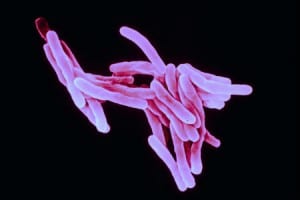
MTB via Flickr
What is tuberculosis
Tuberculosis (TB) is an infectious disease caused by the bacterium Mycobacterium tuberculosis (MTB) that spread via aerosol and primarily affects the lungs.
Although there are current health scares about antibiotic-resistant strains of TB, due to modern sanitation, vaccination programmes and antibiotics there have not been any major TB epidemics in the UK in the 21st century. However, it is currently estimated that one third of the world’s population is infected with various strains of MTB. These infections often pass under the radar because the majority of them are latent, meaning that the infected person does not have any symptoms of the disease.
TB is one of the world’s oldest diseases, in part due to this high level of latency. There are multiple strains of MTB, all associated with differing areas of the globe. What is striking about these strains is that people with TB generally carry the strain of MTB associated with their ethnic origin, despite their current location.
For Dr Donoghue, this is evidence that MTB has evolved with humans. She argued that in the Neolithic and Palaeolithic periods, when humans lived in small populations, pathogens that were highly infectious and killed their hosts quickly failed to survive as they would simply wipe out tribes. In contrast, MTB’s combination of high latency rates and virulence means that carriers transmit the disease before dying.
What’s more, evolving with humans has meant that there are numerous strains of extinct MTB, as well as extant MTB. The research conducted by Dr Donoghue and her team means that new methods are being perfected to analyse these extinct, ancient strains.
How do you analyse extinct bacterium strains?
There are three main methods of identifying historic bacterium strains. The first and oldest method is morphological analysis or, in layman’s terms, studying a person’s bones to look for abnormalities and life-sustained injuries.
The second method, one used in a number of British research projects, is biomarker analysis. Biomarker analysis focuses on proteins and lipids specific to certain pathogens to prove the presence of that pathogen.
Finally, one can use aDNA analysis, a method developed at UCL in the 1990s and the technique preferred by Dr Donoghue. Even though the DNA found on archaeological digs may be thousands of years old and highly fragmented, DNA can often still be sequenced to replicate extinct bacterium, both to confirm its presence and to enable further examination.
What does aDNA tell us about MTB and the people it affected?

Dr Granville’s drawing of Lady Irtyersenu’s mummy
This technology allows researchers to discover much more than previously possible about the deaths and diseases of archaeological skeletons and mummies. One example is Dr Donoghue’s analysis of a 2,500 year old mummy, Lady Irtyersenu.
Dr Granville’s drawing of Lady Irtyersenu’s mummy Lady Irtyersenu was owned by the Regency physician Dr Granville, and in 1825 Dr Granville took it upon himself to conduct an autopsy on Irtyersenu’s mummy to establish her cause of death. A gynaecologist himself, Granville diagnosed Irtyersenu with ovarian cancer.
It wasn’t until 2009 that Granville’s diagnosis was proven incorrect. Dr Donoghue’s team used tissue from the mummy’s lungs, ovaries, gall bladder and bones and found that the ovarian cysts identified by Granville were, in fact, benign. The tissue, however, did show biomarkers of MTB, a diagnosis that was confirmed by aDNA analysis.
With the case of Lady Irtyersenu, Dr Donoghue and her team diagnosed TB as the cause of death; however, in many ancient TB cases MTB is a co-infection, with the host dying from a combination of multiple pathogens, just as the combination of TB and HIV is a large cause of death in the modern world.
This was the case with one of Dr Donoghue’s most recent research projects; a once in a lifetime opportunity to combine fresh archaeological skeletons with rich archival information to shed light on the boom in TB epidemics in 18th century Europe.
In 1994, the bodies of 263 mummies who had died between 1731 and 1838 were discovered in Vác, Hungary, while a church was undergoing renovations, complete with their names, ages and date of deaths written on their coffins.
aDNA anaylsis showed that 89% of the mummies, ranging from newborns to a 95 year old woman, had TB at some point in their lives, and 35% had TB at the time of death, although for some amongst this group their TB was latent.
What was most striking for the researchers was the number of strains of MTB present in the mummies in the crypt. Alongside many co-infections with other pathogens, many of the mummies had two or even three strains of MTB at their time of death, something which was only discoverable through the use of whole genome sequencing, which allowed the researchers to find ‘unknown unknowns’: strains that they did not know existed.
How does analysing aDNA help our understanding of modern disease?
Such breakthroughs are interesting in themselves; however, they also provide pointers for modern researchers. For example, they prove that TB co-infections or multiple strains cannot solely be the fault of antibiotics, since they existed prior to modern medicine.
Moreover, Dr Donoghue’s aDNA sequencing has also proven that MTB has become less aggressive over time, in line with its historic high virulence and high latency rates.
However, for Dr Donoghue this research is clearly a labour of love rather than purely a strive towards medical advancement.
 Close
Close

