Every living cell, whether solitary or part of a larger multicellular organism, is an extremely complex system, involving a multitude of simultaneous chemical reactions regulated by proteins and RNA. Keeping this machine running relies upon a careful balance of gene expression and protein degradation, and cells must be prepared to modulate these processes in response to environmental variation (both internal and external). Homeostatic mechanisms can be an efficient way to regulate gene expression, however one key mechanism – negative autoregulation – is rarely used in organisms like flies and humans. Mathematical modelling by GEE’s Max Reuter and Andrew Pomiankowski shows that evolutionary constraints in the evolution of negative feedback may exist for species who carry multiple copies of each chromosome in their cells.


A Eukaryotic (Animal) Cell and a Prokaryotic (Bacterial) Cell
Homeostasis: From Radiators to Cells
One of the simplest ways to regulate any system is through negative feedback, whereby a particular process is inhibited by the products of that process. This is the system employed by central-heating systems, which use the temperature to regulate the action of a radiator. The radiator pumps out heat, and when the thermometer detects too much heat in the room, it signals the radiator to stop. Negative feedback systems can be highly efficient, stable and responsive.
Negative feedback, or homeostasis, would also be a sensible way to regulate the expression of genes within a cell – a particular gene would continue to be expressed and produce its protein product until there is enough of that product in the cell, at which point expression would stop. This system would be sensitive to the demands of the cell – if the product was being used up quickly, then expression would continue as long as demand was high. In fact, many simple, single-celled organisms, such as E. coli, use this system to respond quickly to changes in their environment.
Around half of all genes in Escherichia coli are regulated by this kind of negative feedback loop, known as negative autoregulation. However, when we look at other organisms such as yeast (Saccharoymyces cerevisiae), fruit flies (Drosophila melanogaster), and humans, we find a very different picture – almost no genes show signs of negative autoregulation (around 2%). If negative autoregulation is such a neat solution to apparently common problem, why aren’t humans and flies using it?
A Matter of Ploidy
One of the key differences between humans and
E. coli (although probably not the one that springs to mind!), is that
E. coli carry only a single copy of each gene in each cell. They are haploid, with a single circular chromosome in each cell that carries a single copy of each gene in the
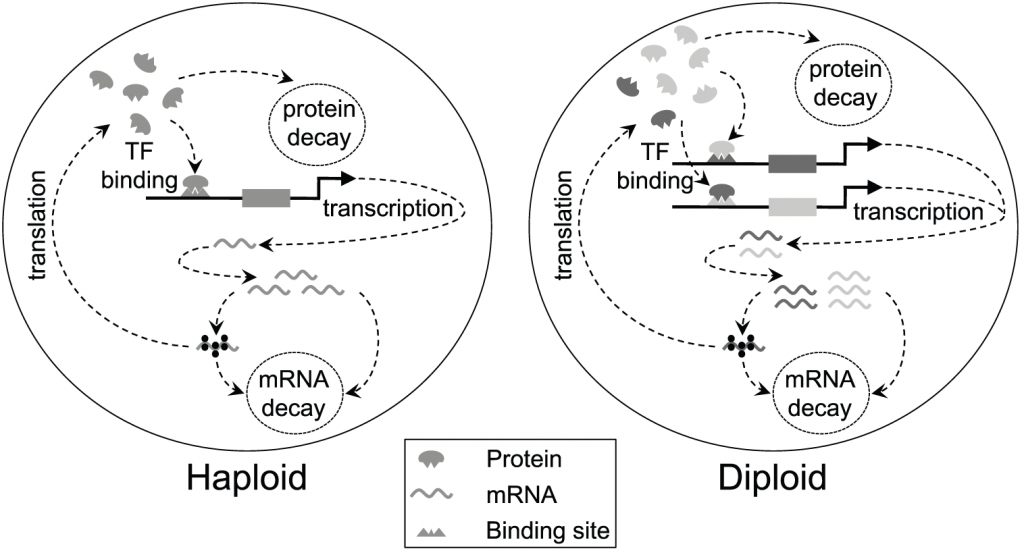
E. coli genome. By contrast, humans, yeast and fruit flies are all diploid, meaning that they carry two copies of each gene in every cell. Our genes are split up into many, straight chromosomes, and we have two copies of each chromosome. Recent research in the department of GEE has used a mathematical modelling approach to investigate how diploidy (having two sets of chromosomes) might constrain the evolution of negative autoregulation.
In a paper in PLoS Computational Biology in March this year, Dr Alexander Stewart, Professor Rob Seymour, Professor Andrew Pomiankowski and Dr Max Reuter from UCL’s GEE produced a mathematical model of how gene regulation might evolve differently in species with one or two sets of chromosomes. Their model focuses on mutations in the promoters of genes, which alter how other protein molecules interact with and repress the expression of those genes.
In haploids, with a single copy of each gene, negative autoregulation produces very tight regulation of gene expression, giving a very rapid response to changing demand. Likewise, in a diploid species with two identical copies of a particular gene, negative autoregulation tends to be beneficial and achieves very efficient gene regulation.
Constraints in the Evolution of Regulation
The problem arises when the diploid carries two different variants of the same gene. This would be the situation whenever a new mutation arises – new mutations appear in a single copy of a particular gene. For a new mutation to be favoured and spread through the population, it must be able to do well, at least initially, as a single copy. The mutated gene must be able to work well alongside the original version. And this is where the problem arises. Stewart, Seymour, Pomiankowski and Reuter (2013) found that mutations that altered the negative autoregulation of a gene didn’t tend to play well with others. Their model considered mutations that alter the strength of the binding site – essentially how strongly regulated that gene is. An individual carrying one strongly regulated gene and one weakly regulated gene actually did worse than an individual with two weakly regulated genes. These heterozygote individuals responded more slowly to changes in demand, and there was more noise in the system. This situation, known as underdominance, where a genetic variant has a lower fitness in the heterozygote form, could be a major constraint to evolution.
Underdominance in negatively autoregulated systems arises because of the disparity in binding site strength between the two different copies of a gene. As each gene pumps out gene product, the stronger binding site is quickly suppressed by the product produced by both genes. It takes much longer for the weaker binding site to be suppressed, as it requires more product to be activated, and most of this is being used up by the stronger binding site. Compared to a haploid, the strong site shows faster response times but the weak site shows a much slower response time, and this averages out to an overall slower response.
Stewart et al (2013)’s model showed that the extent of underdominance depended on how different the two genetic variants were. Large differences in the strength of their binding sites reduced response time and created more noise than smaller differences. Slower response times and increased noise in heterozygotes mean that the maximum strength of regulation achievable in a diploid may be as much as ten-fold lower than in haploids.
Because very small differences between genetic variants in their binding site strength did not experience such a strong effect of underdominance, they were more likely to lead to the evolution of autoregulation. Evolution in diploids could proceed through many very small changes, however there is also likely to be lower limit on the size of mutations – very small changes cannot be ‘seen’ by natural selection and are unlikely to spread. Likewise, multiple binding sites, each relatively weak but which act together cooperatively, were also more likely to overcome the issue of underdominance in the model. However, in general, diploids had a much harder time evolving negative feedback as a mechanism for gene regulation. This evolutionary constraint might have forced diploids such as fruit flies and humans to develop alternative mechanisms to achieve rapid responses, such as increased rates of protein degradation or alternative regulatory mechanisms.
Original Article:


This research was made possible by funding from the Natural Environment Research Council (NERC), the Engineering and Physical Sciences Research Council (EPSRC), and the McDonnell Foundation
 The huge diversity of placental mammals on Earth today first appeared shortly after the mass extinction event that killed the dinosaurs. It is thought that the loss of the dinosaurs, along with much of life on Earth, freed up niches which placental mammals to evolved to fill. But were early placental mammals present, waiting in the wings, during the age of the dinosaurs, or did they appear rapidly after their demise? One recent study suggested that, based on fossil evidence, the placental mammals must have appeared after the cretaceous-tertiary boundary (KT) when dinosaurs and most life on Earth was wiped out. However, a recent paper by GEE’s Mario dos Reis and Ziheng Yang, in collaboration with Philip Donoghue from the University of Bristol, highlights flaws in the methods used in this study, and utilitsed a more thorough approach to show that early placental mammals likely predated the KT boundary.
The huge diversity of placental mammals on Earth today first appeared shortly after the mass extinction event that killed the dinosaurs. It is thought that the loss of the dinosaurs, along with much of life on Earth, freed up niches which placental mammals to evolved to fill. But were early placental mammals present, waiting in the wings, during the age of the dinosaurs, or did they appear rapidly after their demise? One recent study suggested that, based on fossil evidence, the placental mammals must have appeared after the cretaceous-tertiary boundary (KT) when dinosaurs and most life on Earth was wiped out. However, a recent paper by GEE’s Mario dos Reis and Ziheng Yang, in collaboration with Philip Donoghue from the University of Bristol, highlights flaws in the methods used in this study, and utilitsed a more thorough approach to show that early placental mammals likely predated the KT boundary.
 Close
Close