The Challenge of Monitoring Biodiversity
By Claire Asher, on 4 August 2015
a guest blog by Charlie Outhwaite, written for the 2015 Write About Research Competition.
Biological diversity, or biodiversity, is a complex term encompassing the variety of life found on Earth. It incorporates not only differences between species but within species themselves and of the environments and ecosystems where they are found. We as humans benefit a great deal from the biodiversity on Earth in a range of ways; from the clean air we breathe to food, materials and medicines that are produced as a result. These products or services are known as ecosystem services and these services depend on biodiversity. Monitoring the status of biodiversity is therefore an important area of research, but offers its own challenges. New methods offer the chance to utilise data that has been underused in the past due to its associated biases and we are now able to explore and monitor the responses of biodiversity over time for many more species than has previously been possible. This has opened the door not only to more knowledge on a greater range of species but also allows us to look into what aspects are influencing these changes, such as the impact of climate change.
In April 1992, an agreement was signed by a number of government parties to the Convention on Biological Diversity (CBD) agreeing to the global target “to achieve by 2010 a significant reduction of the current rate of biodiversity loss”. Unfortunately, this target was never achieved and so, in 2010 an updated plan was established at the tenth meeting of the Convention in Nagoya, Japan. This revised plan includes 20 main targets, known as the Aichi Biodiversity targets, under 5 strategic goals each encompassing one aspect to benefit biodiversity.
In order to measure progress towards these targets, at both a national and a global scale, a number of indicators of change have been developed, these are often simple graphs showing increases or decreases in the variable being monitored. For the UK, these are published annually by DEFRA (the Department for Environment, Food and Rural Affairs) in the Biodiversity Indicators in your Pocket report (BIYP). Indicators are composite measures of change and are a simple and easy way to communicate change over time. The most recent BIYP report includes a suite of indicators aimed to report on UK progress towards the Aichi targets. These range from indicators of change in volunteer time spent in conservation organisations to assess progress towards strategic goal A (Address the underlying causes of biodiversity loss by mainstreaming biodiversity across government and society) to indicators of the status of UK priority species for Strategic Goal C (To improve the status of biodiversity by safeguarding ecosystems, species and genetic diversity).
However, the monitoring of aspects of the goals is not simple and biodiversity itself provides a great challenge. Take target 12 for example; “By 2020 the extinction of known threatened species has been prevented and their conservation status, particularly of those most in decline, has been improved and sustained”. In order to assess whether the decline and extinction of threatened species has been prevented we need to be able to measure how many there are in the first place, and how that changes over time. Ideally, we would like to go out and count exactly how many there are of each species, but of course this is not possible. It would be difficult enough going out and counting every species in your own garden, let alone across the whole country. So, we have to use the next best alternative. In some cases, standardised monitoring schemes such as the Breeding Bird Survey are set up and species numbers are monitored using standardised techniques across specific sites. This data can then be used to accurately estimate the abundance of those species observed. However, this kind of data is costly to collect and requires a lot of time and effort and so, is not available for the majority of species.
An alternative form of data is biological records. Biological records are a data type that is high in quantity but has a number problems associated with it. Often collected by volunteers through citizen science projects, this type of data can be highly variable in its level of accuracy and completeness. However, with interest and participation in citizen science increasing, the amount of biological records data available is rising. With so much data on hand, and often for those groups of species that are less well studied (such as insects) and for which monitoring scheme data is unavailable, it is important that these data are put to good use. However, because of the problems associated with this data type, it is underused and underappreciated. The main problem is that it is collected in an unstandardized way, which introduces bias into the data. Records will often be collected by an individual at a location of their choosing and they may not report every species they see.
A number of robust statistical methods have been developed that are able to account for these associated biases. Bayesian occupancy models are a complex statistical technique which has been shown by Isaac et al (2015) to most effectively account for the biases of this type of data and produce reliable indicators of change. It is now being used to monitor changes in the biodiversity of less well studied groups of species using biological records from various recording schemes. For these species groups, this kind of data is all that is available and so employing these new methods for analysing biological records is enabling greater research into areas where little is currently known.
However, with human induced drivers being the biggest threat to biodiversity loss, it is not enough to simply monitor changes in species trends. There is a growing need to understand what is causing these trends and how a species’ traits can increase its susceptibility towards these drivers. Through a more thorough understanding of the effects drivers such as climate change have on a group of species, and which species within that group will be most affected, it would be possible to design conservation interventions to target those species most at risk, preventing future declines. This process could act as a form of triage, in determining those species that will be most affected so that conservation and policy action can be targeted to those areas in the first instance. This is becoming increasingly urgent as a mid-term report on progress towards meeting the CBD 2020 targets by Tittensor et al indicates that progress is not positive.
References
- Defra (2014) UK Biodiversity Indicators 2014: Measuring progress towards halting biodiversity loss. Retrieved from http://jncc.defra.gov.uk/page-4229
- Isaac, N. J. B., van Strien, A. J., August, T. A., de Zeeuw, M. P., & Roy, D. B. (2014). Statistics for citizen science: extracting signals of change from noisy ecological data. Methods in Ecology and Evolution, doi:10.1111/2041-210X.12254
- Tittensor, D. P., Walpole, M., Hill, S. L. L., Boyce, D. G., Britten, G. L., Burgess, N. D., … Parks, B. C. (2014). A mid-term analysis of progress toward international biodiversity targets, (October), 1–8.
Charlie is a first year PhD Student based at the Centre for Ecology and Hydrology, Wallingford and working within the Biological Records Centre. Charlie’s PhD is linked with CBER UCL and the RSPB through a CASE partnership. Her research is looking into biodiversity status, drivers and indicators from biological records. Charlie’s interest in measuring and reporting changes in biodiversity has grown since working as an intern and research assistant within the Indicators and Assessments Unit at the Institute of Zoology. Within these roles she worked on the Living Planet Index and on developing a Canadian biodiversity indicator. Going from the reporting and development side of indicators she now hopes to reveal the role of drivers of change and how these interact with species traits to affect changes in biodiversity.
 Close
Close



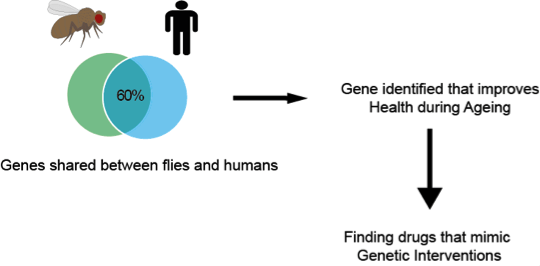
 Jorge graduated with a Medical degree from the Autonomous University of Yucatan, Mexico. After this he completed an MSc in Clinical Neuroscience at the UCL Institute of Neurology, and a PhD in Genetics, Neuroscience and Biogerontology from the UCL Institute of Healthy Ageing (IHA). Currently he works as a Research Associate at the UCL IHA and Research Department of Genetics, Evolution and Environment.
Jorge graduated with a Medical degree from the Autonomous University of Yucatan, Mexico. After this he completed an MSc in Clinical Neuroscience at the UCL Institute of Neurology, and a PhD in Genetics, Neuroscience and Biogerontology from the UCL Institute of Healthy Ageing (IHA). Currently he works as a Research Associate at the UCL IHA and Research Department of Genetics, Evolution and Environment. Box 1. This tree represents the relationship between 5 gene sequences. Each node of the tree either represents a speciation (S1 and S2) or duplication event (star). Thus to know the relation between pairs of genes, you just have to trace them back to their shared node (closest common ancestral copy). In this example, the blue genes between dog and human are orthologous to each other (because they trace back to a speciation event). The red dog and red human genes are also orthologous to each other. However, all the blue genes are paralogous to all the red genes because they trace back to a duplication node. All of these red and blue genes are orthologous to the black (frog) gene, an example of a many:1 relationship.
Box 1. This tree represents the relationship between 5 gene sequences. Each node of the tree either represents a speciation (S1 and S2) or duplication event (star). Thus to know the relation between pairs of genes, you just have to trace them back to their shared node (closest common ancestral copy). In this example, the blue genes between dog and human are orthologous to each other (because they trace back to a speciation event). The red dog and red human genes are also orthologous to each other. However, all the blue genes are paralogous to all the red genes because they trace back to a duplication node. All of these red and blue genes are orthologous to the black (frog) gene, an example of a many:1 relationship. Natasha Glover received her Bachelor of Science and PhD from the Department of Crop and Soil Environmental Science at Virginia Tech in the U.S. Her PhD was focused on plant genomics and biotechnology. She received a Marie Curie International Incoming Fellowship for her first postdoc and worked in Clermont-Ferrand, France at the Institut Nationale de la Recherche Agronomique for 3 years. There, she concentrated on computational biology, with a focus on synteny and duplication in the wheat genome. Natasha is a currently a postdoc based at Bayer CropScience in Ghent, Belgium as part of the Marie Curie PLANT FELLOWS program. Her co-advisor is Dr. Christophe Dessimoz in the department of Genetics, Evolution, and Environment at UCL.
Natasha Glover received her Bachelor of Science and PhD from the Department of Crop and Soil Environmental Science at Virginia Tech in the U.S. Her PhD was focused on plant genomics and biotechnology. She received a Marie Curie International Incoming Fellowship for her first postdoc and worked in Clermont-Ferrand, France at the Institut Nationale de la Recherche Agronomique for 3 years. There, she concentrated on computational biology, with a focus on synteny and duplication in the wheat genome. Natasha is a currently a postdoc based at Bayer CropScience in Ghent, Belgium as part of the Marie Curie PLANT FELLOWS program. Her co-advisor is Dr. Christophe Dessimoz in the department of Genetics, Evolution, and Environment at UCL.